Integrating multiple scRNA-seq data¶
This tutorial shows loading, preprocessing, VIPCCA integration and visualization of 293T and Jurkat cells in three different batches (Mixed Cell Lines).
Importing vipcca package¶
Here, we’ll import vipcca along with other popular packages.
[1]:
import vipcca.model.vipcca as vip
import vipcca.tools.utils as tl
import vipcca.tools.plotting as pl
import matplotlib
import scanpy as sc
matplotlib.use('TkAgg')
# Command for Jupyter notebooks only
%matplotlib inline
import warnings
warnings.filterwarnings('ignore')
from matplotlib.axes._axes import _log as matplotlib_axes_logger
matplotlib_axes_logger.setLevel('ERROR')
Using TensorFlow backend.
Loading data¶
This tutorial uses Mixed Cell Line datasets from 10xgenomics with non-overlapping populations from three batches, two of which contain 293t (2885 cells) and jurkat (3258 cells) cells respectively, and the third batch contains a 1:1 mixture of 293t and jurkat cells (3388 cells).
Read from 10x mtx file The file in 10x mtx format can be downloaded here. Set the fmt parameter of pp.read_sc_data() function to ‘10x_mtx’ to read the data downloaded from 10XGenomics. If the file downloaded from 10XGenomics is in h5 format, the dataset can be loaded by setting the fmt parameter to ‘10x_h5’.
[2]:
base_path = "./data/vipcca/mixed_cell_lines/"
file1 = base_path+"293t/hg19/"
file2 = base_path+"jurkat/hg19/"
file3 = base_path+"mixed/hg19/"
adata_b1 = tl.read_sc_data(file1, fmt='10x_mtx', batch_name="293t")
adata_b2 = tl.read_sc_data(file2, fmt='10x_mtx', batch_name="jurkat")
adata_b3 = tl.read_sc_data(file3, fmt='10x_mtx', batch_name="mixed")
Read from h5ad file The h5ad file we generated that containing the cell type can be downloaded here. Here, we load the three datasets separately.
[3]:
base_path = "./data/vipcca/mixed_cell_lines/"
adata_b1 = tl.read_sc_data(base_path+"293t.h5ad", batch_name="293t")
adata_b2 = tl.read_sc_data(base_path+"jurkat.h5ad", batch_name="jurkat")
adata_b3 = tl.read_sc_data(base_path+"mixed.h5ad", batch_name="mixed")
Each dataset is loading into an AnnData object.
[4]:
adata_b3
[4]:
AnnData object with n_obs × n_vars = 3388 × 32738
obs: '_batch', 'celltype'
var: 'gene_ids'
Data preprocessing¶
Here, we filter and normalize each data separately and concatenate them into one AnnData object. For more details, please check the preprocessing API.
[5]:
adata_all = tl.preprocessing([adata_b1, adata_b2, adata_b3], index_unique="-")
Trying to set attribute `.obs` of view, copying.
Trying to set attribute `.obs` of view, copying.
Trying to set attribute `.obs` of view, copying.
VIPCCA Integration¶
[6]:
# Command for Jupyter notebooks only
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "2"
# The four hyperparameters epochs, lambda_regulizer, batch_input_size, batch_input_size2 are user-defined parameters.
# Given a dataset with ~10K cells, we suggest to pick up
# a value larger than 600 for epochs,
# a value in the range of [2,10] for lambda_regulizer,
# a value at least less than the number of input features for batch_input_size,
# a value in the range of [8,16] for batch_input_size2.
handle = vip.VIPCCA(
adata_all=adata_all,
res_path='./data/vipcca/mixed_cell_lines/',
mode='CVAE',
split_by="_batch",
epochs=20,
lambda_regulizer=5,
batch_input_size=128,
batch_input_size2=16
)
# do integration and return an AnnData object
adata_integrate = handle.fit_integrate()
WARNING:tensorflow:From /Users/zhongyuanke/anaconda3/envs/test-vipcca/lib/python3.7/site-packages/tensorflow_core/python/ops/resource_variable_ops.py:1630: calling BaseResourceVariable.__init__ (from tensorflow.python.ops.resource_variable_ops) with constraint is deprecated and will be removed in a future version.
Instructions for updating:
If using Keras pass *_constraint arguments to layers.
Model: "vae_mlp"
__________________________________________________________________________________________________
Layer (type) Output Shape Param # Connected to
==================================================================================================
encoder_input (InputLayer) (None, 2000) 0
__________________________________________________________________________________________________
batch_input1 (InputLayer) (None, 128) 0
__________________________________________________________________________________________________
encoder_mlp (Model) [(None, 16), (None, 318368 encoder_input[0][0]
batch_input1[0][0]
__________________________________________________________________________________________________
batch_input2 (InputLayer) (None, 16) 0
__________________________________________________________________________________________________
decoder_mlp (Model) (None, 2000) 546272 encoder_mlp[1][2]
batch_input2[0][0]
==================================================================================================
Total params: 864,640
Trainable params: 862,496
Non-trainable params: 2,144
__________________________________________________________________________________________________
WARNING:tensorflow:From /Users/zhongyuanke/anaconda3/envs/test-vipcca/lib/python3.7/site-packages/tensorflow_core/python/ops/math_grad.py:1424: where (from tensorflow.python.ops.array_ops) is deprecated and will be removed in a future version.
Instructions for updating:
Use tf.where in 2.0, which has the same broadcast rule as np.where
WARNING:tensorflow:From /Users/zhongyuanke/anaconda3/envs/test-vipcca/lib/python3.7/site-packages/keras/backend/tensorflow_backend.py:422: The name tf.global_variables is deprecated. Please use tf.compat.v1.global_variables instead.
WARNING:tensorflow:From /Users/zhongyuanke/anaconda3/envs/test-vipcca/lib/python3.7/site-packages/keras/callbacks/tensorboard_v1.py:200: The name tf.summary.merge_all is deprecated. Please use tf.compat.v1.summary.merge_all instead.
WARNING:tensorflow:From /Users/zhongyuanke/anaconda3/envs/test-vipcca/lib/python3.7/site-packages/keras/callbacks/tensorboard_v1.py:203: The name tf.summary.FileWriter is deprecated. Please use tf.compat.v1.summary.FileWriter instead.
Epoch 1/20
9530/9530 [==============================] - 4s 382us/step - loss: 2928.3410
WARNING:tensorflow:From /Users/zhongyuanke/anaconda3/envs/test-vipcca/lib/python3.7/site-packages/keras/callbacks/tensorboard_v1.py:343: The name tf.Summary is deprecated. Please use tf.compat.v1.Summary instead.
Epoch 2/20
9530/9530 [==============================] - 1s 112us/step - loss: 2569.9079
Epoch 3/20
9530/9530 [==============================] - 1s 109us/step - loss: 2555.9494
Epoch 4/20
9530/9530 [==============================] - 1s 108us/step - loss: 2535.0038
Epoch 5/20
9530/9530 [==============================] - 1s 108us/step - loss: 2528.3331
Epoch 6/20
9530/9530 [==============================] - 1s 105us/step - loss: 2520.0686
Epoch 7/20
9530/9530 [==============================] - 1s 106us/step - loss: 2516.1301
Epoch 8/20
9530/9530 [==============================] - 1s 106us/step - loss: 2508.4249
Epoch 9/20
9530/9530 [==============================] - 1s 108us/step - loss: 2500.8367
Epoch 10/20
9530/9530 [==============================] - 1s 107us/step - loss: 2494.5631
Epoch 11/20
9530/9530 [==============================] - 1s 108us/step - loss: 2491.3073
Epoch 12/20
9530/9530 [==============================] - 1s 108us/step - loss: 2486.6252
Epoch 13/20
9530/9530 [==============================] - 1s 110us/step - loss: 2480.4250
Epoch 14/20
9530/9530 [==============================] - 1s 109us/step - loss: 2480.4351
Epoch 15/20
9530/9530 [==============================] - 1s 113us/step - loss: 2480.8911
Epoch 16/20
9530/9530 [==============================] - 1s 115us/step - loss: 2476.3571
Epoch 17/20
9530/9530 [==============================] - 1s 114us/step - loss: 2475.4372
Epoch 18/20
9530/9530 [==============================] - 1s 114us/step - loss: 2472.8237
Epoch 19/20
9530/9530 [==============================] - 1s 115us/step - loss: 2470.7086
Epoch 20/20
9530/9530 [==============================] - 1s 115us/step - loss: 2468.2385
... storing '_batch' as categorical
... storing 'celltype' as categorical
[7]:
adata_integrate
[7]:
AnnData object with n_obs × n_vars = 9530 × 2000
obs: '_batch', 'celltype', 'n_genes', 'percent_mito', 'n_counts', 'size_factor', 'batch'
var: 'gene_ids', 'n_cells-0-0', 'highly_variable-0-0', 'means-0-0', 'dispersions-0-0', 'dispersions_norm-0-0', 'n_cells-1-0', 'highly_variable-1-0', 'means-1-0', 'dispersions-1-0', 'dispersions_norm-1-0', 'n_cells-1', 'highly_variable-1', 'means-1', 'dispersions-1', 'dispersions_norm-1'
obsm: 'X_vipcca'
1.The meta.data of each cell has been saved in adata.obs
2.The embedding representation from vipcca of each cell have been saved in adata.obsm(‘X_vipcca’)
Loading result¶
Loading result from saved model.h5 file through vipcca: The model.h5 file of the trained result can be downloaded here
[8]:
model_path = 'model.h5'
handle = vip.VIPCCA(
adata_all=adata_all,
res_path='./data/vipcca/mixed_cell_lines/',
mode='CVAE',
split_by="_batch",
epochs=20,
lambda_regulizer=5,
batch_input_size=128,
batch_input_size2=16,
model_file=model_path
)
adata_integrate = handle.fit_integrate()
Model: "vae_mlp"
__________________________________________________________________________________________________
Layer (type) Output Shape Param # Connected to
==================================================================================================
encoder_input (InputLayer) (None, 2000) 0
__________________________________________________________________________________________________
batch_input1 (InputLayer) (None, 128) 0
__________________________________________________________________________________________________
encoder_mlp (Model) [(None, 16), (None, 318368 encoder_input[0][0]
batch_input1[0][0]
__________________________________________________________________________________________________
batch_input2 (InputLayer) (None, 16) 0
__________________________________________________________________________________________________
decoder_mlp (Model) (None, 2000) 546272 encoder_mlp[1][2]
batch_input2[0][0]
==================================================================================================
Total params: 864,640
Trainable params: 862,496
Non-trainable params: 2,144
__________________________________________________________________________________________________
... storing '_batch' as categorical
... storing 'celltype' as categorical
Loading result from h5ad file: The output.h5ad file of the trained result can be downloaded here
[9]:
integrate_path = './data/vipcca/mixed_cell_line/output.h5ad'
adata_integrate = sc.read_h5ad(integrate_path)
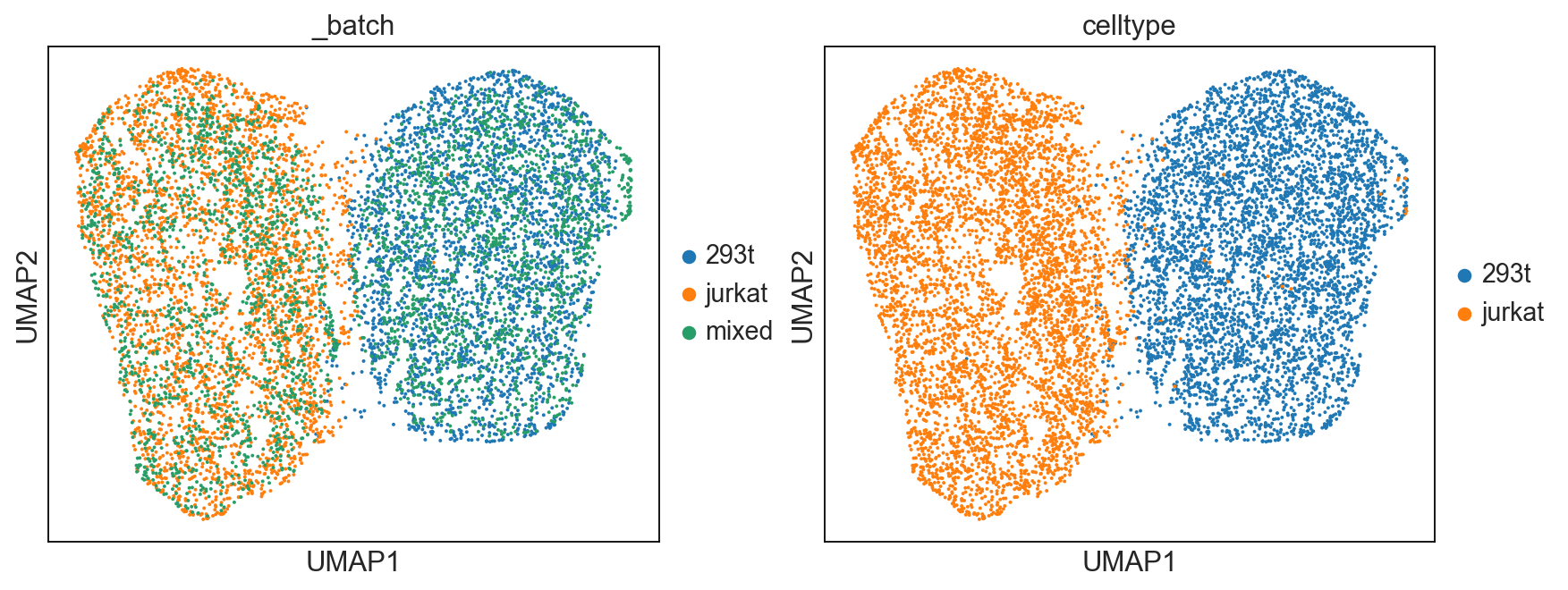
UMAP Visualization¶
We use UMAP to reduce the embedding feature output by vipcca in 2 dimensions.
[8]:
sc.pp.neighbors(adata_integrate, use_rep='X_vipcca')
sc.tl.umap(adata_integrate)
Visualization of UMAP result.
[9]:
sc.set_figure_params(figsize=[5.5,4.5])
sc.pl.umap(adata_integrate, color=['_batch','celltype'], use_raw=False, show=True,)
... storing '_batch' as categorical
... storing 'celltype' as categorical
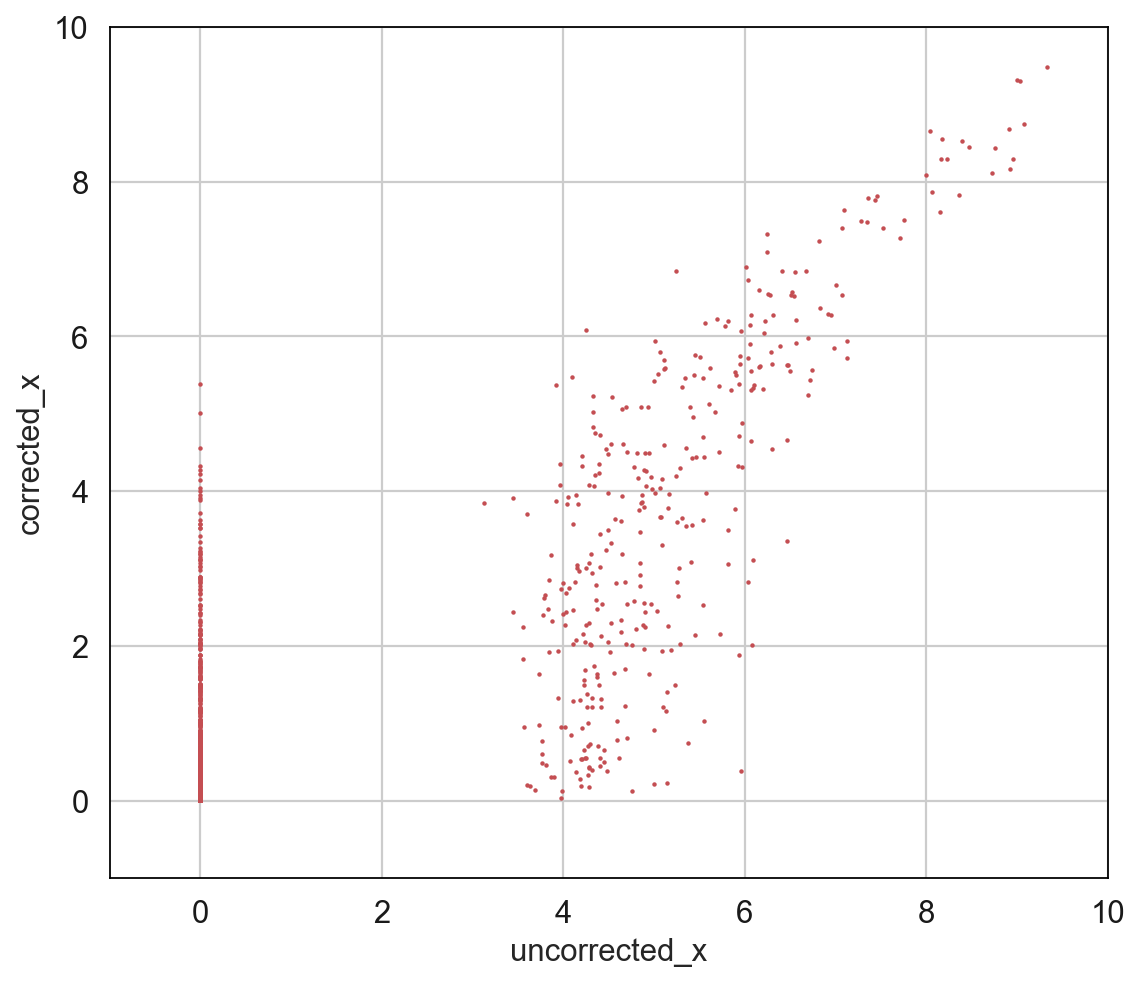
Plot correlation¶
This function is only applicable to AnnData generated by fit_integrate() function training. Randomly select 2000 locations in the gene expression matrix. The x-axis represents the expression value of the original data at these locations, and the y-axis represents the expression value of the data after vipcca integration at the same location.
[10]:
pl.plotCorrelation(adata_integrate.raw.X, adata_integrate.X, save=False, rnum=2000, lim=10)